Running USM3D (Legacy)
Memory requirements and usage of USM3D (Legacy)
The base version of USM3D requires 45 words (360 bytes) of memory per cell for an explicit scheme and 179 words (1432 bytes) of memory per cell for an implicit scheme. Although, implicit scheme requires more memory it is recommended as it is more robust for both inviscid and viscous calculations. While it is theoretically possible to perform viscous calculations using the explicit scheme, it has not been well tested and calibrated. Therefore, use of explicit scheme, if inevitable, is recommended only for inviscid calculations.
The flow solver is available to the eligible users in the object code form. The source code has been compiled on several different computer platforms to build suitable machine code. The executables have been built to perform double precision calculations for the following platforms, using FORTRAN 77 compiler unless stated otherwise.
| Platform | Name of the executable |
|---|---|
| CRAY T932 (FORTRAN 90) | usm3d_52.t90 |
| CRAY C-90 (FORTRAN 90) | usm3d_52.c90 |
| SGI R12000/R10000 | usm3d_52.sgi |
| SGI R12000/R10000 (FORTRAN 90) | usm3d_52.sgif90 |
| SUN UltraSPARC(200 MHz) | usm3d_52.sun |
| PC cluster (DEC Alpha 21264/500 MHz) | usm3d_52.linux |
Before beginning a run for a specific case, a user can make certain diagnostic checks pertaining to the availability of the adequate memory on the host machine and required files. The run-time memory required by the flow solver can be estimated by the following command.
The variable proj_name has to be suitably replaced by the identifying case name.
usm3d_52.xxx -s proj_name
OR
usm3d_52.xxx -S proj_name
A check for the existence of all the files required for the run can be made by the following command.
usm3d_52.xxx -c proj_name
OR
usm3d_52.xxx -C proj_name
Note that the check above is specifically designed for supporting files such as, proj_name.mapbc file and proj_name.fandm files. Other required files proj_name.cogsg and proj_name.iface must exist for this check to work satisfactorily.
To initiate a run that stores the flowfield output (for flow visualization) and a flow restart solution in files with the name proj_name.flo and proj_name.urest respectively, the following command should be given.
usm3d_52.xxx proj_name
However, in certain situations such as a parametric study, it may be desired to direct the flowfield output and restart solution to respective files with a prefix different from that of grid files, namely, proj_name. In such situations, the following command should be given to initiate a run.
usm3d_52.xxx proj_name flo_name
Performance on different platforms
The CPU performance of the flow solver has been assessed on several compute platforms by considering ONERA M6 wing geometry in a transonic turbulent separated flow environment. A fully viscous tetrahedral unstructured grid that resolves the viscous sublayer has been used for this purpose. The grid consists of 495,012 tetrahedral cells, 86,389 nodes, and 12,986 boundary faces. This case has been run on several computer platforms to evaluate the the time required by the solver on a per cell per iteration basis. For all the runs, the numerical solution control parameters have been held constant. The runs have been made for 1500 iterations using implicit scheme. The solver requires about 179 words (1432 bytes) of memory per cell for an implicit, viscous run in double precision. The accuracy and the convergence characteristics of the solver for this case and several other cases have been evaluated in the next chapter.
| Platform | µsec/cell/iteration | MFLOPS |
|---|---|---|
| CRAY T932 | 16.78 | 487 |
| CRAY C-90 | 28.69 | 286.13 |
| SGI R12000 | 171.90 | 47.76 |
| SGI R10000 | 221.24 | 37.10 |
| SUN UltraSPARC(200 MHz) | 341.13 | 24.06 |
| PC cluster (DEC Alpha 21264/500 MHz) | 97.76 | 83.96 |
Output files
All the output files generated during a run have been summarized below. The various files and their formats have been described following the table.
| File name | Purpose | When created? | Update frequency |
|---|---|---|---|
| proj_name.flo | Solution post-processing | ipltqn > 0 | end of a run |
| proj_name.urest | Solution restart | Always | every nwrest, end of a run |
| hist.plt | Convergence monitoring | Always | every iteration |
| tet.out | input echo, grid statistics, forces and moments with respect to various axis systems and components, CPU time and memory information |
Always | every iteration |
| temp.flo | Interim solution investigation | When “1” is placed in usm3d.write file |
iteration at which code finds “1” in the usm3d.write file |
| cells.1storder | Diagnostics: identification of cells with pressure < pmin |
idiagnos > = 0 and if any cell pressure < pmin occurs |
every iteration after pressure < pmin occurs for any cell |
| temp.vist | Diagnostics: field visualization of turbulent viscosity, turbulent to laminar viscosity and Spalart-Allmaras turbulence model variable |
idiagnos > = 0 and ivisc > = 2, |
intermediate solution write using usm3d.write file, end of a run |
| temp.cfyplus |
Diagnostics: skin-friction distribution on viscous surface for analytically generated unstructured grid(e.g. flat plate) |
idiagnos <= 2 and ivisc > 0 | intermediate solution write using usm3d.write file, end of a run |
| info.diag |
Diagnostics for minimum pressure, maximum turbulent viscosity, average Y+ |
idiagnos > 0 | every iteration |
| cells.problem | Diagnostics: identification of cells with mass flux residue 100 times the corresponding average over the domain |
idiagnos > 0 | every iteration |
| resmax.plt |
Diagnostics: field visualization of unit volume stagnation energy residual extrema |
idiagnos > 1 | end of a run |
| temp.cfyplus* |
Diagnostics: skin-friction distribution on viscous surface for VGRID-based unstructured grid |
idiagnos > 2 and ivisc > 0 | intermediate solution write using usm3d.write file, end of a run |
| yplus.plt | variation of Y+(turbulent case) or normalized cartesian velocities, u and w(laminar case) across the boundary layer |
nodeypl > 0 and ivisc > 0 |
intermediate solution write using usm3d.write file, end of a run |
This file contains flowfield data for each grid node in the form of conserved fluid-dynamic quantities, namely, the density (![]() ), x-momentum component (
), x-momentum component (![]() ), y-momentum component (
), y-momentum component (![]() ), z-momentum component (
), z-momentum component (![]() ), and total energy (
), and total energy (![]() ). The file can be either unformatted (ipltqn = 1) or formatted (ipltqn = 2).
). The file can be either unformatted (ipltqn = 1) or formatted (ipltqn = 2).
The unformatted file is written by:
write(ifile)xmach
write(ifile)((qn(i,j),i=1,nnode),j=1,5)
whereas, the formatted file is written by:
write(ifile,*)xmach
do 30 i=1,nnode
write(ifile,*)i,(qn(i,j),j=1,5)
30 continue
where:
| xmach | = | freestream Mach number |
|---|---|---|
| nnode | = | total number of nodes (points) in the grid |
| qn(i,1..5) | = | an array containing a vector of conserved fluid dynamic quantities for each node |
This is an unformatted file. The following code snippet describes the file format.
write(lrf) version
write(lrf) xmach,ncell,nnode,nface,nbndr,nfbnd,nt,iordflg,
. nswitch
write(lrf) ((q(nc,l),nc=1,ncell),l=1,5),
. ((qb(nf,l), nf=1,nfbnd),l=1,5),
. ((qgh(nf,l),nf=1,nfbnd),l=1,5),(rsid(n),n=1,nt),
. (cll(n),n=1,nt),(cdd(n),n=1,nt),(cdv(n),n=1,nt),
. (cmm(n),n=1,nt),(ctim(n),n=1,nt),(cfl(n),n=1,nt),
. (rmstnu(n),n=1,nt),(cf(n),n=1,nbndr),
. (factor(nc),nc=1,ncell),(dtv(nc),nc=1,ncell),
. (pintavg(n),n=1,4)
if(ivisc.gt.1)
.write(lrf) (tnu(nc),nc=1,ncell),(smag(nc),nc=1,ncell)
write(lrf) bcoef,sc,cfll,ntss,ntsm,bflag,icntlim,alogres,
. cfl1
where:
| version | = | code version number |
|---|---|---|
| xmach | = | freestream Mach number |
| ncell | = | total number of tetrahedral cells in the grid |
| nnode | = | total number of nodes (points) in the grid |
| nface | = | total number of faces (triangular) in the grid |
| nbndr | = | number of boundary nodes in the grid |
| nfbnd | = | number of boundary faces (triangular) in the grid |
| nt | = | total number of iterations since the fresh run for a case |
| iordflg | = | flag indicating the global order of accuracy for the spatial discretization |
| nswitch | = | iteration at which the spatial discretization accuracy changed from first to second order (when iorder = 0) |
| q(i,1..5) | = | an array containing a vector of conserved fluid dynamic quantities for each tetrahedral cell |
| qb(i,1..5) | = | an array containing a vector of conserved fluid dynamic quantities for each boundary face |
| rsid | = | L2-norm of the mean flow residue * ncell, at each iteration |
| cll,cdd,cdv,cmm | = | vectors of aerodynamic total lift, total drag, viscous drag, and total pitching moment respectively, at each iteration |
| ctim | = | cumulative CPU time at each iteration since the fresh run for a case |
| cfl | = | CFL number at each iteration |
| rmstnu | = | L2-norm of the turbulence flux residue * ncell, at each iteration |
| cf | = | a vector of skin-friction-magnitude for each boundary node |
| factor | = | a vector governing local accuracy of the computations for each tetrahedral cell |
| dtv | = | time-step vector for each tetrahedral cell |
| pintavg | = | a vector of average intake pressure for each engine |
| tnu | = | a vector of the primary variable in Spalart-Allmaras turbulence model equation for each tetrahedral cell |
| smag | = | a vector of vorticity magnitude for each tetrahedral cell |
| bcoef,sc,cfll ntss,ntsm,bflag, icntlim,alogres,cfl1 |
= | variables related to various algorithmic implementations inside the code |
The format of a portion of the history file is shown below. Note that the header statements have been omitted for the sake of brevity.
do 35 k=1,nt
write(75,1) k,rsidn,cll(k),cdd(k),cdv(k),cmm(k),cfl(k),rsidt
35 continue
1 format(i8,7(e14.6))
where:
| nt | = | instantaneous total of the number of iterations completed |
|---|---|---|
| rsidn | = | Log10[rsid(k)/rsid(1)], where rsid is the L2-norm of the mean flow residue |
| cll,cdd,cdv,cmm | = | aerodynamic total lift, total drag, viscous drag, and total pitching moment respectively, at each iteration |
| rsidt | = | Log10[rmstnu(k)/rmstnu(1)], where rmstnu is the L2-norm of the turbulence flux residue |
This file contains several strands of information related to a run. Therefore, the representative contents of this file have been shown first.
* * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * *
* * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * *
* * * *
* * USM3D : CHARACTERISTIC-BASED SCHEME FOR STEADY SOLUTIONS TO THE * *
* * EULER/N-S EQUATIONS ON UNSTRUCTURED TETRAHEDRAL MESHES * *
* * * *
* * VERSION 5.2 : Configuration Aerodynamics Branch, MS 499 * *
* * NASA Langley Research Center, Hampton, VA * *
* * January 2000. * *
* * * *
* * * *
* * THIS VERSION IS AN ENHANCEMENT OF THE PREVIOUS VERSION 5.13. * *
* * THE NEW FEATURES DEVELOPED AND IMPLEMENTED BY: * *
* * * *
* * DR. NEAL T. FRINK * *
* * NASA LANGLEY RESEARCH CENTER * *
* * Configurations Aerodynamics Branch, MS 499 * *
* * Aerodynamics Competency * *
* * Hampton, VA 23681-0001 * *
* * phone: (757) 864-2864 * *
* * FAX: (757) 864-8469 * *
* * email: n.t.frink@larc.nasa.gov * *
* * * *
* * AND * *
* * * *
* * DR. MOHAGNA J. PANDYA * *
* * PARAGON RESEARCH ASSOCIATES, INC. * *
* * 28 Research Drive * *
* * Hampton, VA 23666 * *
* * phone: (757) 766-8695 * *
* * FAX: (757) 766-9323 * *
* * email: mpandya@paragon.langley.net * *
* * * *
* * DISCLAIMER: VERSION 5.2 IS STILL UNDER DEVELOPMENT. * *
* * PLEASE REPORT ALL PROBLEMS. * *
* * * *
* * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * *
* * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * * *
1USM3D (v5.2 Export) - Flat plate Turbulent BL Y+node=80.0
Mach alpha beta ReUe Tinf,dR itwall Tw/Tinf ipwall
0.50000 0.00000 0.00000 0.200E+07 518.68800 0 1.05000 1
sref cref bref xmc ymc zmc
1.00000 1.00000 1.00000 0.00000 0.00000 0.00000
impl dt/cfl1 iramp clf2 cflmin GS_tol
1 -10.00 40 200.00 5.00 0.00000
irest mstage iresmth dqmax pbreak pmin limiter
0 3 1 0.25000 0.01000 0.00100 0.00000
nupdate nwrest ipltqn idiagnos nodeypl
10 300 2 2 969
iorder lapl-avg high-bc ifds ivisc ev_lim
0 2 1 1 3 0.100
ncyc nengines compF&M cldes
3000 0 0 0
*******************************
Allocated 16.18 MB of Memory
*******************************
********************************
caution: coloring scheme skipped
********************************
nbcolor= 1
ntcolor= 2
Volume Extrema: Minimum= 0.33181E-08 Cell= 5274
Maximum= 0.39882E-05 Cell= 10086
Initializing "tnu" to 1.341946000000000
Grid characteristics: ncell = 10368
nnode = 2793
nface = 22728
nbndr = 1994
nfbnd = 3984
Automated differencing . . .
First order
Iter RMS resid. LOG(R/R0) CL CM CPU SECS.
Updated time step . . .
1 0.395239E-05 0.000000E+00 -0.217122E-06 0.759414E-07 0.186339E+01
Updated time step . . .
2 0.393131E-05 -0.232263E-02 -0.754503E-06 0.263045E-06 0.360000E+01
Updated time step . . .
3 0.390963E-05 -0.472407E-02 -0.154461E-05 0.534991E-06 0.533616E+01
Updated time step . . .
4 0.388859E-05 -0.706714E-02 -0.242079E-05 0.829564E-06 0.707147E+01
Updated time step . . .
5 0.386778E-05 -0.939723E-02 -0.320627E-05 0.108105E-05 0.880593E+01
Updated time step . . .
6 0.384633E-05 -0.118129E-01 -0.437911E-05 0.144174E-05 0.105545E+02
Updated time step . . .
7 0.381635E-05 -0.152113E-01 -0.510503E-05 0.163347E-05 0.123022E+02
Updated time step . . .
8 0.378103E-05 -0.192490E-01 -0.553025E-05 0.170987E-05 0.140500E+02
Updated time step . . .
9 0.374249E-05 -0.236989E-01 -0.577415E-05 0.171516E-05 0.157976E+02
Updated time step . . .
10 0.370200E-05 -0.284232E-01 -0.592015E-05 0.167974E-05 0.175455E+02
Updated time step . . .
11 0.366064E-05 -0.333022E-01 -0.601178E-05 0.161874E-05 0.192936E+02
Updated time step . . .
12 0.361914E-05 -0.382540E-01 -0.606753E-05 0.153790E-05 0.210430E+02
Updated time step . . .
13 0.357797E-05 -0.432230E-01 -0.609454E-05 0.143901E-05 0.227912E+02
Updated time step . . .
14 0.353743E-05 -0.481716E-01 -0.609671E-05 0.132303E-05 0.245414E+02
Updated time step . . .
15 0.349772E-05 -0.530744E-01 -0.607798E-05 0.119131E-05 0.262900E+02
Updated time step . . .
16 0.345895E-05 -0.579157E-01 -0.604313E-05 0.104576E-05 0.280374E+02
Updated time step . . .
17 0.342116E-05 -0.626860E-01 -0.599763E-05 0.888880E-06 0.297853E+02
Updated time step . . .
18 0.338439E-05 -0.673793E-01 -0.594733E-05 0.723628E-06 0.315326E+02
Updated time step . . .
19 0.334864E-05 -0.719912E-01 -0.589806E-05 0.553283E-06 0.332804E+02
Updated time step . . .
20 0.331392E-05 -0.765179E-01 -0.585525E-05 0.381273E-06 0.350285E+02
21 0.328022E-05 -0.809559E-01 -0.582338E-05 0.210914E-06 0.367272E+02
22 0.324756E-05 -0.853025E-01 -0.580568E-05 0.450908E-07 0.384257E+02
23 0.321591E-05 -0.895559E-01 -0.580415E-05 -0.113944E-06 0.401241E+02
24 0.318525E-05 -0.937154E-01 -0.581971E-05 -0.264720E-06 0.418229E+02
25 0.315558E-05 -0.977808E-01 -0.585242E-05 -0.406644E-06 0.435215E+02
26 0.312685E-05 -0.101752E+00 -0.590140E-05 -0.540048E-06 0.452197E+02
27 0.309904E-05 -0.105632E+00 -0.596469E-05 -0.666143E-06 0.469183E+02
28 0.307210E-05 -0.109425E+00 -0.603928E-05 -0.786790E-06 0.486174E+02
29 0.304598E-05 -0.113133E+00 -0.612149E-05 -0.904041E-06 0.503169E+02
30 0.302064E-05 -0.116760E+00 -0.620797E-05 -0.101972E-05 0.520657E+02
31 0.299606E-05 -0.120309E+00 -0.629483E-05 -0.113498E-05 0.537645E+02
... ... ... ... ... ...
... ... ... ... ... ...
295 0.398805E-06 -0.996099E+00 0.127374E-03 -0.633313E-04 0.503279E+03
296 0.396870E-06 -0.998211E+00 0.127539E-03 -0.634247E-04 0.504976E+03
297 0.394951E-06 -0.100032E+01 0.127703E-03 -0.635182E-04 0.506674E+03
Switching to higher-order differencing . . .
---No limiter applied---
298 0.778195E-05 0.294229E+00 0.177054E-03 -0.815376E-04 0.508408E+03
299 0.326283E-05 -0.832644E-01 0.130151E-03 -0.643720E-04 0.510118E+03
Updated time step . . .
300 0.267567E-05 -0.169428E+00 0.120911E-03 -0.605827E-04 0.511876E+03
Writing version 5.2 restart file for unstructured solution
... ... ... ... ... ...
... ... ... ... ... ...
3000 0.163320E-10 -0.538382E+01 0.114179E-03 -0.637174E-04 0.513941E+04
Writing version 5.2 restart file for unstructured solution
Writing version 5.2 restart file for unstructured solution
FORCES integrated over 136 boundary faces:
(Integrated surface contains 105 nodes.)
--------------------------------------------
B O D Y A X I S C O E F F I C I E N T S:
C_X, C_Y, C_Z, Cm_X, Cm_Y, Cm_Z:
PRESSURE COMPONENT:
0.00000E+00 0.00000E+00 0.11418E-03 -0.22836E-05 -0.63717E-04 0.00000E+00
SKIN-FRICTION COMPONENT:
0.14371E-03 0.16737E-12 0.23593E-22 -0.47185E-24 -0.69158E-23 0.28742E-05
TOTAL COMBINED FORCES
0.14371E-03 0.16737E-12 0.11418E-03 -0.22836E-05 -0.63717E-04 0.28742E-05
------------------------------------------------------
S T A B I L I T Y A X I S C O E F F I C I E N T S:
CLs ,CDs ,CYs , Cpitchs, Crolls, Cyaws:
PRESSURE COMPONENT:
0.11418E-03 0.00000E+00 0.00000E+00 -0.63717E-04 -0.22836E-05 0.00000E+00
SKIN-FRICTION COMPONENT:
0.23593E-22 0.14371E-03 0.16737E-12 -0.69158E-23 -0.47185E-24 0.28742E-05
TOTAL COMBINED FORCES
0.11418E-03 0.14371E-03 0.16737E-12 -0.63717E-04 -0.22836E-05 0.28742E-05
--------------------------------------------
W I N D A X I S C O E F F I C I E N T S:
CLw ,CDw ,CYw , Cpitchw, Crollw, Cyaww:
PRESSURE COMPONENT:
0.11418E-03 0.00000E+00 0.00000E+00 -0.63717E-04 -0.22836E-05 0.00000E+00
SKIN-FRICTION COMPONENT:
0.23593E-22 0.14371E-03 0.16737E-12 -0.69158E-23 -0.47185E-24 0.28742E-05
TOTAL COMBINED FORCES
0.11418E-03 0.14371E-03 0.16737E-12 -0.63717E-04 -0.22836E-05 0.28742E-05
NORMALIZED
CYCLE RESIDUAL RESIDUAL
3000 0.1633E-10 0.4132E-05
DENSITY EXTREMA
Maximum: 0.1003E+01 Cell: 5269
Minimum: 0.9809E+00 Cell: 284
PRESSURE EXTREMA
Maximum: 0.7178E+00 Cell: 5269
Minimum: 0.7121E+00 Cell: 5318
TEMPERATURE EXTREMA
Maximum: 0.1019E+01 Cell: 284
Minimum: 0.9991E+00 Cell: 5893
ENTROPY EXTREMA
Maximum: 0.1026E+01 Cell: 284
Minimum: 0.9997E+00 Cell: 5893
VORTICITY EXTREMA
Maximum: 0.2083E+03 Cell: 5322
Minimum: 0.1404E-05 Cell: 9510
OVERHEAD TIME : 0.4434E+00 sec.
TOTAL CYCLE TIME: 0.5139E+04 sec.
TIME PER CYCLE : 0.1713E+01 sec.
TIME PER CELL : 0.1652E-03 sec.
TOTAL MEMORY: 2128328 words
Words/Cell : 205
CPU TIME = 0.5140E+04
Note that the contents correspond to a fresh run (starting from the freestream initialization). For this case, the quantities such as flow residue, aerodynamic lift, drag and pitching moment etc. are written on this file at every iteration. For a restart run, this file provides the snapshots of the variation of these quantities in previous runs of this case by detailing their values for the first 19 iterations and subsequently for every iteration in the multiple of the hundred. The format statement for writing these quantities is shown below.
write(lw,100) nt,rms,alogres,cll(nt),cmm(nt),ctim(nt)
100 format(i5,5e15.6)
where:
| nt | = | instantaneous total of the number of iterations completed |
|---|---|---|
| rsid | = | L2-norm of the mean flow residue * ncell |
| rsidn | = | Log10[rsid(k)/rsid(1)], |
| cll,cmm | = | aerodynamic total lift, and total pitching moment respectively, at each iteration |
| ctim | = | Total CPU time consumed until the current iteration |
The output and the format of this file are identical to that of a proj_name.flo file.
USM3Dns searches for the cells whose pressure has fallen below a user-defined pressure level pmin and tags them. The search process is performed at every iteration and if the tagged cells are found, the cell index and the corresponding cell centered pressure value of such cells are written on the presently considered file. To improve the robustness, USM3Dns locally reduces the tagged cells to first-order spatial accuracy while maintaining the overall global computations with higher-order spatial accuracy. These tagged cells remain tagged until a solution is restarted with pmin < 0. This file is written with the statements shown below.
if(iorder.eq.1)then
WRITE(88,*) '1st-ORDER CELLS WITH P<PMIN=',pminimum
else if(iorder.eq.2)then
WRITE(88,*) 'CELLS WITH P<PMIN=',pminimum,
. ' REDUCED TO 1st ORDER IN FDS'
end if
do 600 i=1,ncell
if(factor(i).eq.0) WRITE(88,888) i, q(i,5)
600 continue
888 format(i10, e20.5)
where:
| iorder | = | flag that controls the global accuracy of the computations |
|---|---|---|
| pminimum | = | user-specified minimum pressure threshold, pmin |
| ncell | = | total number of tetrahedral cells in the grid |
| factor | = | a vector governing local accuracy of the computations for each tetrahedral cell |
| q(i,5) | = | local pressure at a tetrahedral cell center |
This is a formatted file that is written out only for a viscous turbulent case. It mimics the structure of a flow file that has last five columns (columns 2-6) that contain the values of density, x-, y-, z-momentum components and the total energy. However, in the present file the three momentum components (columns 3-5) are substituted with the three quantities, namely, turbulent viscosity, ratio of turbulent to laminar viscosity, and the primary variable of the Spalart-Allmaras turbulence model. Whereas, columns 2 and 6 (columns containing the density and total energy in the flow file) both contain a variable set to unity. The specific structure of the file facilitates use of any post-processing code in analyzing the variations of the turbulence quantities mentioned above, provided that these quantities are treated as cartesian velocity components while using the post-processing code. The format of this file is as shown below.
write(77,*) xmach
do 6 i=1,nnode
write(77,777) i,rone,vist,chifv1,tnun(i),rone
777 format(i8, f5.0, 3e15.7, f5.0)
where:
| xmach | = | freestream Mach number |
|---|---|---|
| nnode | = | total number of nodes (points) in the grid |
| rone | = | 1. |
| vist | = | turbulent viscosity( |
| chifv1 | = | ratio of turbulent-to-laminar viscosity ( |
| tnun | = | primary variable of Spalart-Allmaras model ( |
This file contains information for all the grid nodes on the viscous patches (so called viscous nodes) only. The file format is shown below.
write(78,*)'# X, Y, Z, CF, YPLUS'
do 8 i=1,ntwo
write(78,178) nb,xnd(nb),ynd(nb),znd(nb),cfx,yplus(nb)
8 continue
178 format(i8,3e14.6,2e15.7)
where:
| ntwo | = | total number of viscous nodes |
|---|---|---|
| xnd,ynd,znd | = | cartesian coordinates triple at a viscous node |
| cfx | = | x-component of the local skin-friction coefficient at a viscous node |
| yplus | = | Y+ of the very first node on the normal drawn from a viscous surface node |
The format of a portion of this file is shown below. Note that the file header statements have been omitted for the sake of brevity.
if(ivisc .eq. 0 .or. ivisc .eq. 1)then
write(81,4) nt,cpmin,icpmin,nfirst
4 format(i6,2x,e11.4,i10,2x,i10)
else
write(81,1) nt,cpmin,icpmin,nfirst,vmax,ayplusn, ayplusc
1 format(i6,2x,e11.4,i10,2x,i10,4x,e11.5,2f8.1)
endif
where:
| nt | = | current iteration number |
|---|---|---|
| cpmin | = | minimum cell-centered pressure in an entire computational domain at any iteration |
| icpmin | = | index of the cell with the minimum pressure at any iteration |
| nfirst | = | total number of tagged cells whose pressure has fallen below a user-specified minimum pressure threshold, pmin, at any instance during a run. |
| vmax | = | maximum cell-centered turbulent viscosity ( at any iteration |
| ayplusn | = | average Y+of all the immediate grid nodes off a viscous surface |
| ayplusc | = | average Y+of all the cells around a viscous surface |
Note that the last two quantities above are computed only for a wall function -based flow analysis. For a case that fully resolves the sub-layer (full viscous grid), these quantities are not computed by the code presently, and the output file will print their values as zero.
This file lists all the cells whose mass flux residual magnitude (per unit cell volume) is two-orders-of-magnitude greater than the corresponding average over the entire computational domain. These cells can be plotted with a graphic analysis tool such as ViGPLOT or FAST, to identify a trouble spot in a computational domain.
write(88,*)'CELLS WITH NORMALIZED RESIDUAL ERROR GREATER',
. ' THAN 100-TIMES THE AVERAGE, cycle=',nt
write(88,*)'Cell log(r/r_avg)'
do 30 i=1,ncell
if(rcell.ge.2.) write(88,601) i, rcell
30 continue
601 format(i10,f10.3)
where:
| ncell | = | total number of tetrahedral cells in the grid |
|---|---|---|
| rcell | = | 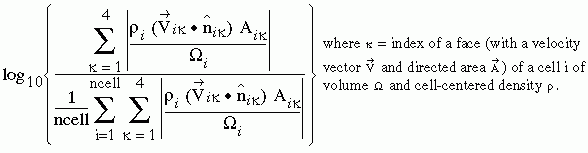 |
| rcell | = | index of a cell with rcell >= 2 |
This is a formatted file. It mimics the structure of a flow file that has last five columns (columns 2-6) that contain the values of density, x-, y-, z-momentum components and the total energy. However, in the present file the three momentum components (columns 3-5) are substituted with the three quantities, namely, absolute maximum, maximum and minimum values of the cell total energy residual error per unit volume. These extrema for a node are chosen locally from all the cells surrounding that specific node as shown in the sketch below.
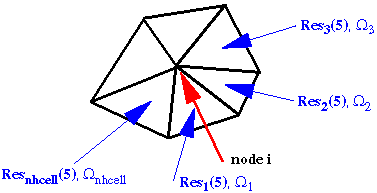
Whereas, columns 2 and 6 (columns containing the density and total energy in the flow file) both contain a variable set to unity. The specific structure of the file facilitates use of any post-processing code in analyzing the variations of the quantities mentioned above. For example, one can plot the iso-surface of “u-velocity” and visually locate the pockets of large residual error. The format of this file is as shown below.
write(79,*) xmach
do 63 i=1,nnode
63 write(79,630) i,rone,resE1,resE2,resE3,rone
630 format(i8, f8.2, 3e15.7, f8.2)
where:
| xmach | = | freestream Mach number |
|---|---|---|
| nnode | = | total number of nodes (points) in the grid |
| rone | = | 1. |
| resE1 | = | 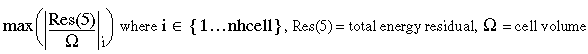 |
| resE2 | = |  |
| resE3 | = |  |
This is a formatted file. It mimics the structure of a flow file that has last five columns (columns 2-6) that contain the values
of density, x-, y-, z-momentum components and the total energy. However, in the present file the three momentum components (columns 3-5) are substituted with the three quantities, namely, skin-friction coefficient in both x (streamwise) and y (spanwise) directions as well as Y+ of the node that is immediately adjacent to the surface node under consideration and lies on the normal drawn from it. Note that these quantities are relevant only for the surface nodes lying on the viscous patches. The corresponding quantities for interior nodes have been fixed to an arbitrarily chosen values and written on the file to constitute a flow-like file. One can study the skin-friction distribution on any arbitrary cross-section of the geometry by using the post-processing code, usgutil. The format of this file is as shown below.
write(78,*) xmach
do 98 n = 1,nnode
write(78,*) n,rone,cfx,cfy,ypl,rone
98 continue
where:
| xmach | = | freestream Mach number |
|---|---|---|
| nnode | = | total number of nodes (points) in the grid |
| cfx,cfy | = | x and y components respectively, of local skin-friction coefficient for a surface node (arbitrary value of 1000 for an interior node) |
| ypl | = | Y+ of the node that is immediate adjacent to the surface node n and on lies on the normal drawn from it. (arbitrary value of 1000 for an interior node) rone = 1. |
This file provides a distribution of certain boundary layer quantities of interest along a normal drawn from the viscous surface node, nodeypl. The specific quantities written out on this file (illustrated below) depend on whether a run corresponds to a laminar or a turbulent flow. The file output is mainly useful for accessing the accuracy of the solver and adequacy of a grid for simpler geometries such as a flat plate boundary layer. The format of a portion of this file is shown below. Note that the file header statements have been omitted for the sake of brevity.
do 40 i=2,20
WRITE(74,200) yp,up,dplus,qm,n1
40 continue
200 format(4e15.7, i10)
where, for a turbulent flow analysis case:
| n1 | = | grid node index of an interior node inside the boundary layer that lies on a normal drawn from the viscous surface node, nodeypl |
|---|---|---|
| yp,up | = | Y+ and U+ respectively, of the node under consideration |
| dplus | = | distance (dimensional) of the node under consideration from the viscous surface node, nodeypl |
| qm | = | Velocity magnitude at the node under consideration |
and for a laminar flow analysis case:
| n1 | = | grid node index of an interior node inside the boundary layer that lies on a normal drawn from the viscous surface node, nodeypl |
|---|---|---|
| yp | = | non-dimensional distance 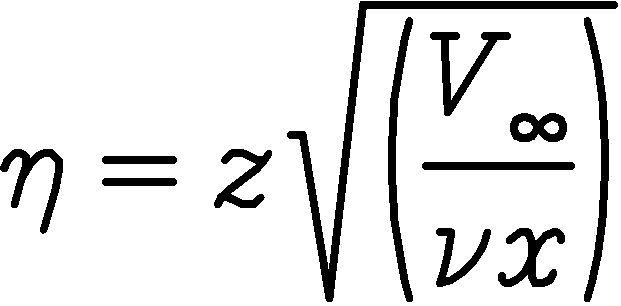 , of the node under , of the node underconsideration from the viscous surface node, nodeypl |
| up,dplus | = | |
| qm | = | Velocity magnitude, consideration |
Special features
Compatibility of proj_name.mapbc and proj_name.inpt files:
At the beginning of the run, the flow solver automatically checks for the consistency between the flow analysis option, ivisc selected and the type of boundary condition specified for the boundary patches. Therefore, if a user has selected inviscid flow analysis option (ivisc = 0) but the proj_name.mapbc file contains a patch with a viscous boundary condition (boundary condition type 4), then the flow solver detects this inconsistency and terminates the run and writes a message on tet.out fileas shown below.
write(lw,*)’STOP: “.mapbc” file not compatible ‘
write(lw,*)’Change appropriate wall BC to inviscid’
The flow solver also alerts a user if it finds that the user has selected viscous flow analysis option (ivisc > 0) but the proj_name.mapbc file contains a patch with an inviscid boundary condition (boundary condition type 5). However, it does not terminate the run. The warning message is written out on the tet.out file prior to initiating the time integration loop and it is shown below.
write(lw,*)’WARNING: “.mapbc” file may not be compatible ‘
write(lw,*)’May have inviscid b.c. when you want viscous
Converged solution for a desired lift:
The flow solver has a feedback algorithm for obtaining a converged flow solution for a desired lift coefficient by perturbing the user specified freestream angle-of-attack. This feature can be invoked by specifying the desired lift coefficient as an input parameter cldes. Additionally, a user is also required to provide the correct values of the reference chord length cref and the reference span (half-span for a half configuration geometry) bref. Before entering the feedback loop, the flow solver uses the input-specified angle-of-attack and stabilizes the flow such that the flow residual errors drop by one-and-a half order of magnitude compared to their initial value. Once in the feedback loop, the flow solver adjusts the angle-of-attack at every iteration to yield a desired lift coefficient within the tolerance of 0.001. The adjustment of the angle-of-attack is based on a lift curve slope:
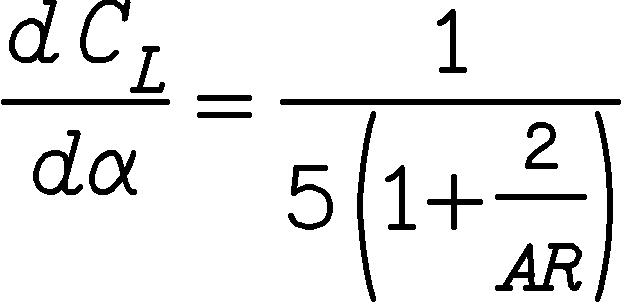 where, AR = Aspect ratio
where, AR = Aspect ratio
WRITE(lw,*) ‘Adjusting alpha to ‘,alpha*57.2958e0
Note that this loop is activated only for a non-zero value of cldes.
Monitoring and stopping the run execution:
The flow solver updates the files, hist.plt and tet.out at every iteration to relay the information related to the case convergence to a user. It also has a provision to prematurely stop a run before the the number of cycles ncyc, specified at the beginning of a run, are completed. Before stopping a run, the flow solver writes all the relevant output files including the solution restart file and the flow visualization file. This feature can be invoked by creating a file named as usm3d.stop with an entry of “1”. The file must be in the same directory from where the run execution command was given. The flow solver checks for the existence of the file and the entry of “1” in it at every iteration and if the check is positive, stops the run immediately. This feature is helpful for a case that may be gradually diverging. In such circumstances, user may want to interrupt the run, analyze the flow results and identify the cause of the convergence difficulty.
Writing output files without stopping run execution:
In addition to interrupting a run using the usm3d.stop file, the flow solver also allows for writing an intermediate output without stopping the run. This feature can be activated by creating a file named as usm3d.write with an entry of “1”. The file must be in the the same directory from where the run execution command was given. The flow solver checks for the existence of the file and the entry of “1” in it at every iteration and if the check is positive, writes out intermediate output. The flow file temp.flo is always created. Other output files such as, temp.vist, temp.cfyplus, and yplus.plt may also be generated subject to the input parameters specified by a user.
